Version 1.1.1 (2019)
Please read the README for details in updates with respect to the previous version.
Please read the REQUIREMENTS page for details in operating system requirements.
Definition
"GeneBase" is a fully structured local
database with a simple graphic interface for personal
computers which allows users to do original calculations and
searches for any information about eukaryotic genes
annotated in the National Center for Biotechnology
Information's (NCBI) Gene database.
GeneBase 1.1.1 Database Design Report
Download
Pre-loaded versions of GeneBase 1.1.1 filled with human data and sequences*:
Macintosh
Windows
Pre-loaded versions of GeneBase 1.1.1 filled with human data (sequences excluded):
Macintosh
Windows
Empty (template) version of GeneBase 1.1.1 with Python scripts for parsing NCBI Gene entries and NCBI Nucleotide sequences included:
Macintosh
Windows
GeneBase 1.1.1 Database Design Report
Download
Pre-loaded versions of GeneBase 1.1.1 filled with human data and sequences*:
Macintosh
Windows
Pre-loaded versions of GeneBase 1.1.1 filled with human data (sequences excluded):
Macintosh
Windows
Empty (template) version of GeneBase 1.1.1 with Python scripts for parsing NCBI Gene entries and NCBI Nucleotide sequences included:
Macintosh
Windows
*Due to the presence of sequences which are indexed in order to improve sequence searches in "Gene_Table", this version is slower than the version with sequences excluded in making summary calculations shown in the correlating "Report" table. Sequences are not necessary to calculate this summary, thus if the user is interested only in gene and exon/intron number and length statistics, the use of the pre-loaded versions of GeneBase 1.1.1 filled with only human data (sequences excluded) is preferable.
Description of the main steps of the analysis
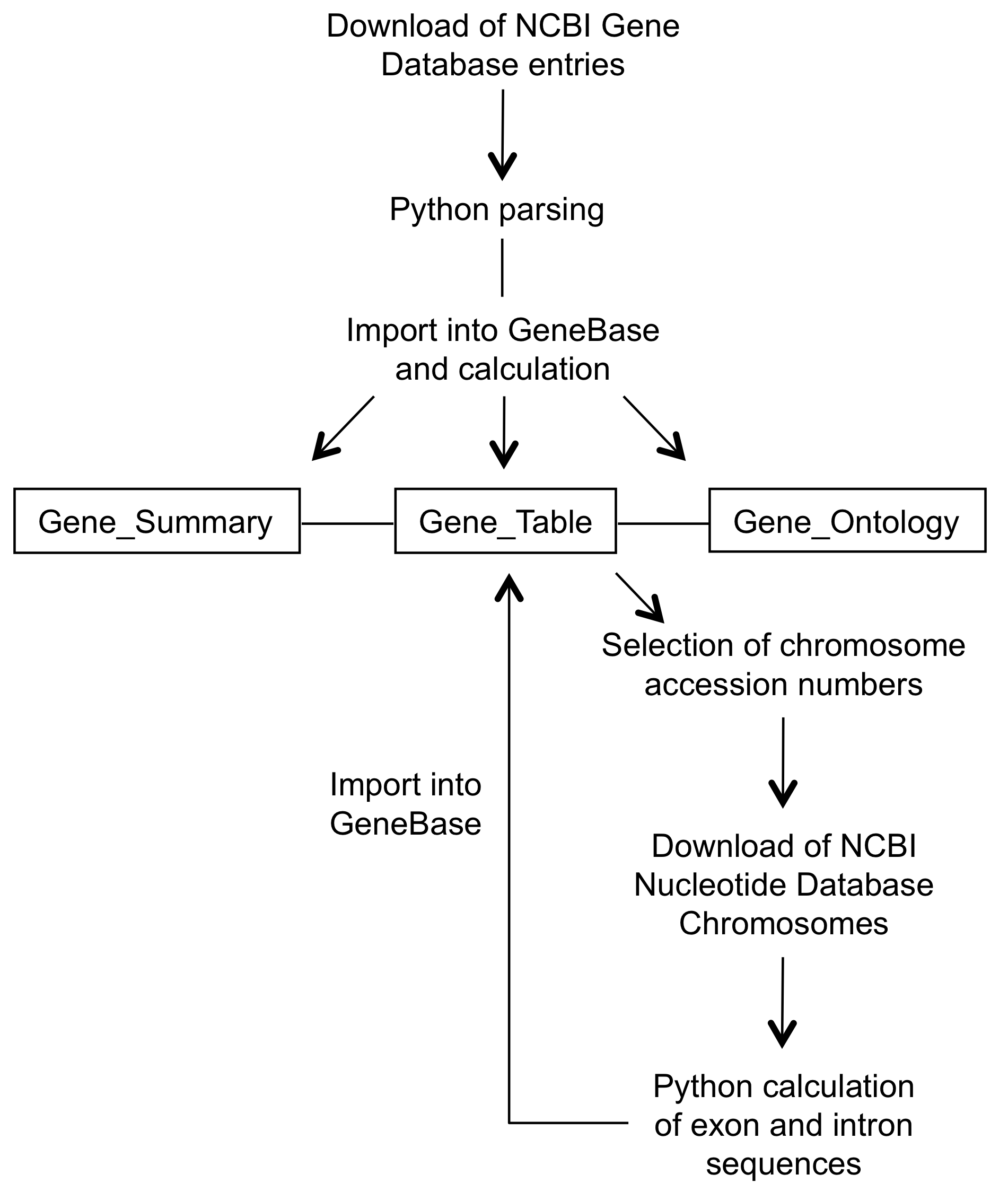
First,
the user is guided to download, parse and import NCBI's
Gene database entries in GeneBase.
GeneBase 1.1.1 contains three correlated tables: "Gene_Summary" collects details about each gene, such as the official gene symbol, the official gene full name, the organism name and a brief description of the gene; "Gene_Table" consists of one record for each exon including the corresponding intron (if an intron follows that exon), representing the exon/intron structure of each transcript isoform; "Gene_Ontology" contains specific Gene Ontology labels, codes and terms for each gene, when available.
In addition, a table named "Reports" is generated to provide statistics such as the mean lengths of exons and introns. A table named "Transcripts" shows a set of useful fields from "Gene_Summary" and "Gene_Table" tables, in order to give an overview of main available information for each transcript. Finally, a table named "Genes" shows a set of useful fields from "Gene_Summary" and "Gene_Table" tables, in order to give an overview of main available information for each gene. Here only the transcript isoform with the highest number of exons is arbitrarily shown for each gene.
Furthermore, following the download of the chromosome sequences from NCBI Nucleotide database, the user can extract and import exon and intron sequences.
GeneBase 1.1.1 contains three correlated tables: "Gene_Summary" collects details about each gene, such as the official gene symbol, the official gene full name, the organism name and a brief description of the gene; "Gene_Table" consists of one record for each exon including the corresponding intron (if an intron follows that exon), representing the exon/intron structure of each transcript isoform; "Gene_Ontology" contains specific Gene Ontology labels, codes and terms for each gene, when available.
In addition, a table named "Reports" is generated to provide statistics such as the mean lengths of exons and introns. A table named "Transcripts" shows a set of useful fields from "Gene_Summary" and "Gene_Table" tables, in order to give an overview of main available information for each transcript. Finally, a table named "Genes" shows a set of useful fields from "Gene_Summary" and "Gene_Table" tables, in order to give an overview of main available information for each gene. Here only the transcript isoform with the highest number of exons is arbitrarily shown for each gene.
Furthermore, following the download of the chromosome sequences from NCBI Nucleotide database, the user can extract and import exon and intron sequences.
Each software table presents a box showing
useful related fields of other related software
tables, giving the opportunity to perform crossed
searches. A sample screenshot of the "Gene_Table" software section representing the
exon/intron structure of each transcript
isoform, with corresponding sequences and
related Gene Ontology categories:
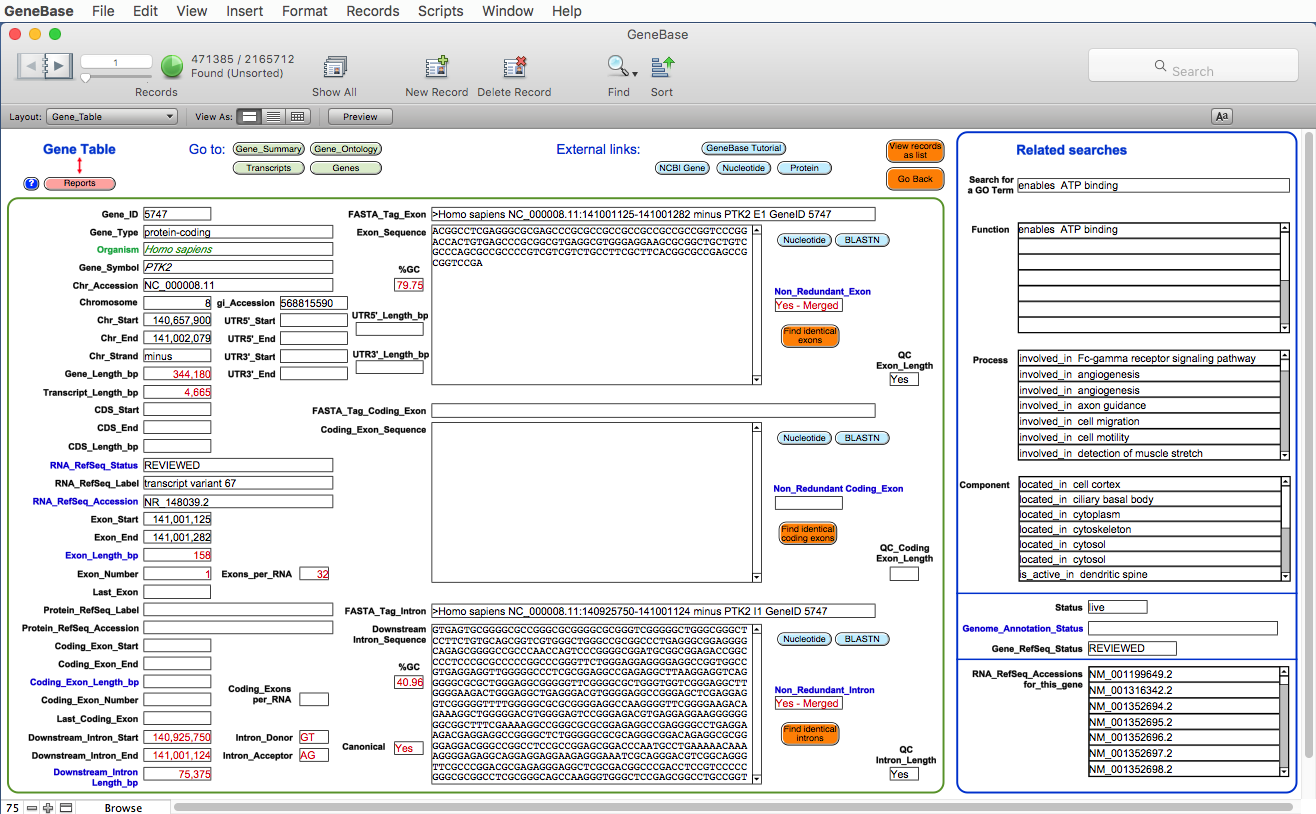
Useful information
specifically calculated by GeneBase 1.1.1, which is not
available in NCBI's Gene database, is highlighted in red.
Tutorial
Tutorial
This Tutorial guides the
user through a step-by-step process in order to set up and
use the software for the analysis of any organism.