Version 1.0 (2015)
Definition
"GeneBase" is a fully structured local
database with a simple graphic interface for personal
computers which allows users to do original calculations and
searches for any information about eukaryotic genes
annotated in the National Center for Biotechnology
Information's (NCBI) Gene database.
Download
Pre-loaded versions of GeneBase filled with eukaryotic data and sequences:
Macintosh
Windows
Pre-loaded versions of GeneBase filled with only eukaryotic data (sequences excluded):
Macintosh
Windows
Empty (template) version of GeneBase with Python scripts for parsing NCBI Gene entries and NCBI Nucleotide sequences included:
Macintosh
Windows
Download
Pre-loaded versions of GeneBase filled with eukaryotic data and sequences:
Macintosh
Windows
Pre-loaded versions of GeneBase filled with only eukaryotic data (sequences excluded):
Macintosh
Windows
Empty (template) version of GeneBase with Python scripts for parsing NCBI Gene entries and NCBI Nucleotide sequences included:
Macintosh
Windows
GeneBase Database Design Report
Description of the main steps of the analysis
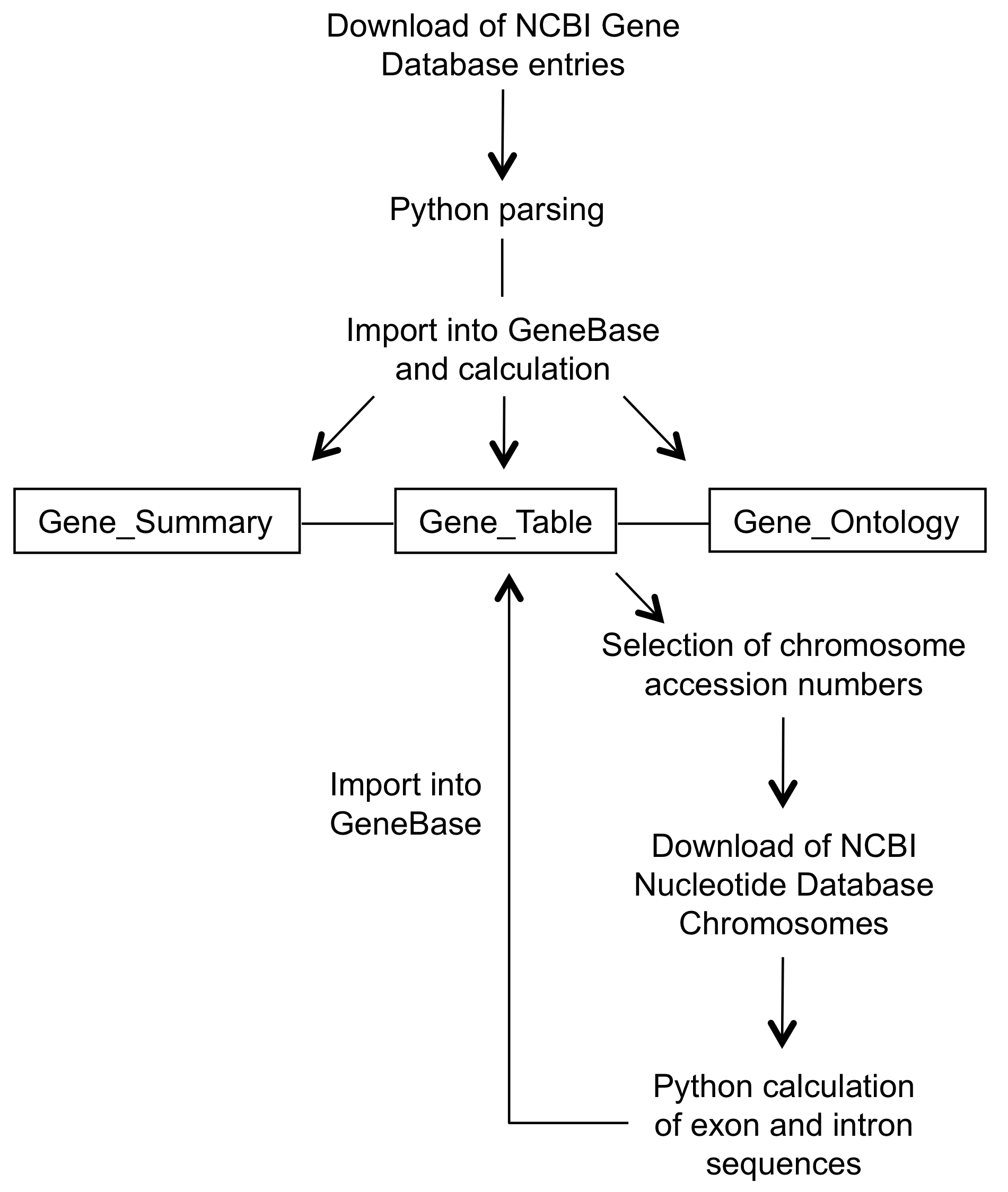
First, the user is guided to download, parse
and import NCBI's Gene database entries in GeneBase.
GeneBase contains three correlated tables: "Gene_Summary" collects details about each gene, such as the official gene symbol, the official gene full name, the organism name and a brief description of the gene; "Gene_Table" consists of one record for each exon including the corresponding intron (if an intron follows that exon), representing the exon/intron structure of each transcript isoform; "Gene_Ontology" contains specific Gene Ontology labels, codes and terms for each gene, when available.
In addition, a table named "Reports" is generated to provide statistics such as the mean lengths of exons and introns.
Furthermore, following the download of the chromosome sequences from NCBI Nucleotide database, the user can extract and import exon and intron sequences.
GeneBase contains three correlated tables: "Gene_Summary" collects details about each gene, such as the official gene symbol, the official gene full name, the organism name and a brief description of the gene; "Gene_Table" consists of one record for each exon including the corresponding intron (if an intron follows that exon), representing the exon/intron structure of each transcript isoform; "Gene_Ontology" contains specific Gene Ontology labels, codes and terms for each gene, when available.
In addition, a table named "Reports" is generated to provide statistics such as the mean lengths of exons and introns.
Furthermore, following the download of the chromosome sequences from NCBI Nucleotide database, the user can extract and import exon and intron sequences.
Each software table presents a box showing
useful related fields of other related software
tables, giving the opportunity to perform crossed
searches. A sample screenshot of the 'Gene_Table' software section representing the
exon/intron structure of each transcript
isoform, with corresponding sequences and
related Gene Ontology categories:
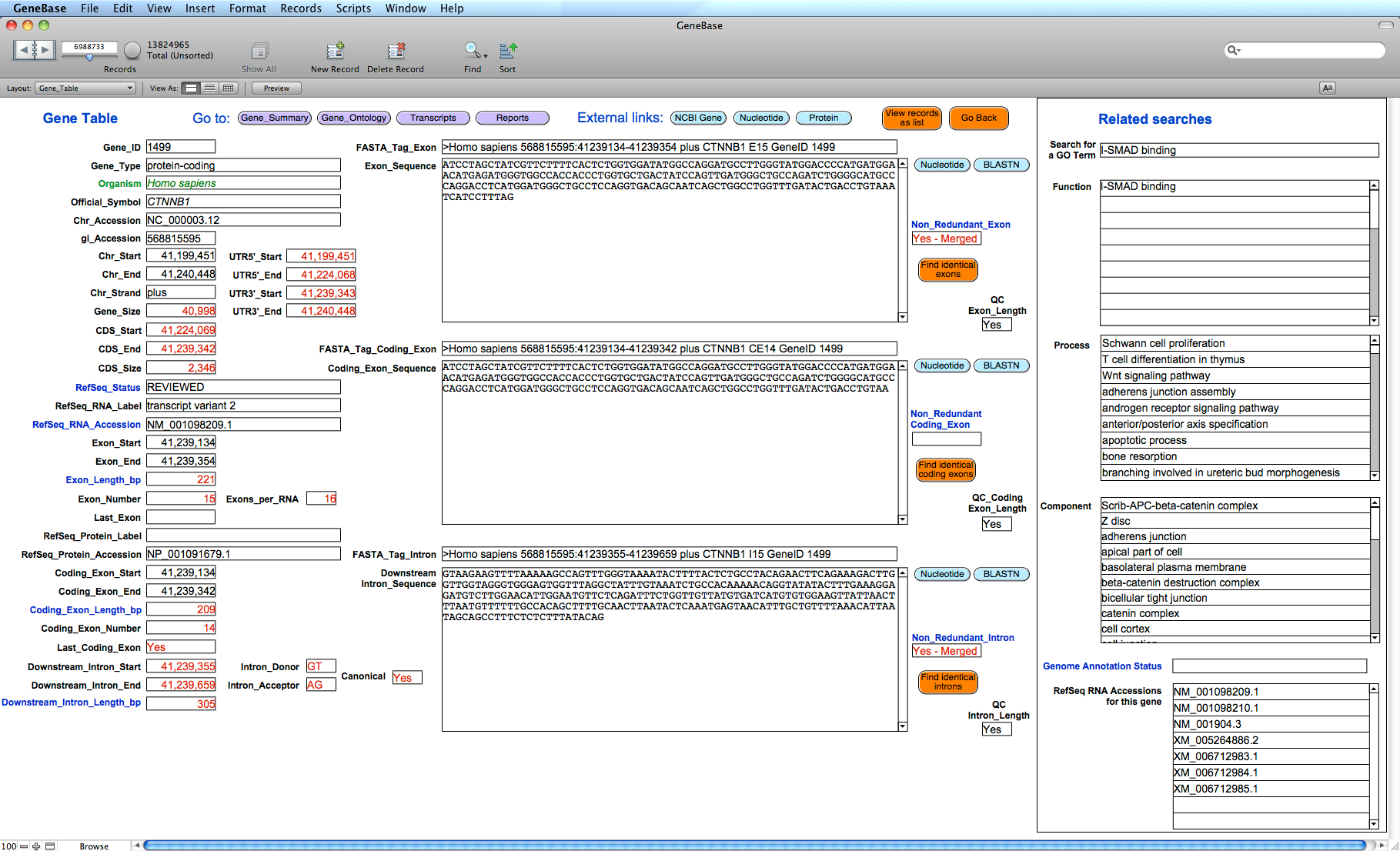
Useful information
specifically calculated by GeneBase, which is not available
in NCBI's Gene database, is highlighted in red.
Database construction
We downloaded from NCBI Gene all current (alive/live) eukaryotic records with a genomic gene source (excluding gene models) available up to April 22nd, 2015. We obtained 679,451 entries for Animalia (Metazoa), 1,203,082 for Fungi and 534,875 for Plants (Viridiplantae), for a total of 359 organisms. Among the 2,417,408 total gene entries, 76,182 are "Reviewed", 41,862 "Validated", 2,245,205 "Provisional", 31,464 "Inferred", 22,691 "Predicted", 1 "Model" and 1 "Withdrawn" (despite the gene model exclusion performed using the web search described in the tutorial).
After the initial parsing and importing steps, the three main tables in GeneBase database are constituted as follows: "Gene_Summary" contains 2,417,408 records (one for each NCBI Gene entry), "Gene_Table" (Figure) contains 13,824,965 records (one record for each gene exon, included the corresponding intron if an intron follows that exon) and "Gene_Ontology" contains 149,064 records in all (one for each gene with GO information available). Due to the lack of annotated transcribed products, a gene structure was not available for 86,824 Gene unique identifiers (UIDs).
Among the total gene entries, 2,368,726 are protein coding, 25,796 pseudogenes (pseudo), 21,247 non coding RNA (ncRNA), 527 coding for small nucleolar RNA (snoRNA), 137 for small nuclear RNA (snRNA), 86 for ribosomal RNA (rRNA) and 6 for cytoplasmic RNA genes (scRNA) (the remaining are not specified).
Then, in order to integrate nucleotide sequences, from the "Gene_Table" table of our database, we selected 861,550 with the "Validated" RefSeq status and 534,578 records with the "Reviewed" RefSeq status (in both cases having an "NM_" or "NR_" type of RefSeq RNA accession number, in order to exclude "XM_" or "XR_" model Refseq records generated by automated pipelines) for a total of 1,396,128 exon entries. Using Batch Entrez we were able to retrieve and download 1,336 records out of the 1,338 corresponding chromosome sequences. This selection gave rise to a total of 1,385,944 "Gene_Table" records, which represent 10% of all available entries, updated with exon, coding exon (for protein coding genes) and the corresponding downstream intron sequences up to May 5th, 2015. The whole database including sequences has a size of 25.1 gigabytes following decompression.
The whole process, including data import and processing, required about 4 days for completion (1 additional day was required to obtain exon and intron sequences).
Tutorial
Database construction
We downloaded from NCBI Gene all current (alive/live) eukaryotic records with a genomic gene source (excluding gene models) available up to April 22nd, 2015. We obtained 679,451 entries for Animalia (Metazoa), 1,203,082 for Fungi and 534,875 for Plants (Viridiplantae), for a total of 359 organisms. Among the 2,417,408 total gene entries, 76,182 are "Reviewed", 41,862 "Validated", 2,245,205 "Provisional", 31,464 "Inferred", 22,691 "Predicted", 1 "Model" and 1 "Withdrawn" (despite the gene model exclusion performed using the web search described in the tutorial).
After the initial parsing and importing steps, the three main tables in GeneBase database are constituted as follows: "Gene_Summary" contains 2,417,408 records (one for each NCBI Gene entry), "Gene_Table" (Figure) contains 13,824,965 records (one record for each gene exon, included the corresponding intron if an intron follows that exon) and "Gene_Ontology" contains 149,064 records in all (one for each gene with GO information available). Due to the lack of annotated transcribed products, a gene structure was not available for 86,824 Gene unique identifiers (UIDs).
Among the total gene entries, 2,368,726 are protein coding, 25,796 pseudogenes (pseudo), 21,247 non coding RNA (ncRNA), 527 coding for small nucleolar RNA (snoRNA), 137 for small nuclear RNA (snRNA), 86 for ribosomal RNA (rRNA) and 6 for cytoplasmic RNA genes (scRNA) (the remaining are not specified).
Then, in order to integrate nucleotide sequences, from the "Gene_Table" table of our database, we selected 861,550 with the "Validated" RefSeq status and 534,578 records with the "Reviewed" RefSeq status (in both cases having an "NM_" or "NR_" type of RefSeq RNA accession number, in order to exclude "XM_" or "XR_" model Refseq records generated by automated pipelines) for a total of 1,396,128 exon entries. Using Batch Entrez we were able to retrieve and download 1,336 records out of the 1,338 corresponding chromosome sequences. This selection gave rise to a total of 1,385,944 "Gene_Table" records, which represent 10% of all available entries, updated with exon, coding exon (for protein coding genes) and the corresponding downstream intron sequences up to May 5th, 2015. The whole database including sequences has a size of 25.1 gigabytes following decompression.
The whole process, including data import and processing, required about 4 days for completion (1 additional day was required to obtain exon and intron sequences).
Tutorial
This Tutorial
guides the user through a step-by-step process in order to
set up and use the software for the analysis of any
organism.