A Python executable script which counts all types of nucleotide letters was developed in order to calculate exact base composition of chromosome sequences. The obtained counts can be imported into a suitable basic database, available on request, developed within the FileMaker Pro Advanced environment in order to perform length and weight calculations. The Python script named "calculate_bases.py" and the FileMaker database named "DNA" are available here:
Macintosh
Windows
Requirements
Minimum software requirements are:
Mac
OS X 10.6, OS X Lion 10.7, OS X Mountain Lion 10.8;
Windows XP Professional, Home Edition (Service Pack 3);
Windows Vista Ultimate, Business, Home Premium (Service Pack 2);
Windows 7 Ultimate, Professional, Home Premium;
Windows 8 Standard and Pro edition.
Minimum system requirements are:
Mac OS X 10.6, Intel-based Mac CPU (Central Processing Unit), 1 GigaByte (GB) of RAM (Random Access Memory), 1024x768 or higher resolution video adapter and display.
Windows XP Professional, Home Edition (Service Pack 3), 700 MegaHertz (MHz) CPU or faster, 256 MegaBytes (MB) of RAM, 1024x768 or higher resolution video adapter and display.
Windows XP Professional, Home Edition (Service Pack 3);
Windows Vista Ultimate, Business, Home Premium (Service Pack 2);
Windows 7 Ultimate, Professional, Home Premium;
Windows 8 Standard and Pro edition.
Minimum system requirements are:
Mac OS X 10.6, Intel-based Mac CPU (Central Processing Unit), 1 GigaByte (GB) of RAM (Random Access Memory), 1024x768 or higher resolution video adapter and display.
Windows XP Professional, Home Edition (Service Pack 3), 700 MegaHertz (MHz) CPU or faster, 256 MegaBytes (MB) of RAM, 1024x768 or higher resolution video adapter and display.
A connection to the Internet
is required to display the software Guide and to
download data for set up, but not to run the tool.
The downloaded file
should be automatically decompressed, generating a
"Base_Counts" folder.
Failing this, double click on the file to activate the default decompression utility of your system.
Failing this, double click on the file to activate the default decompression utility of your system.
The
Base_Counts Folder contains:
- DNA Folder with:
"DNA.app" (Macintosh) or "DNA.exe" (Windows) file
(the runtime application);
"DNA.fmp12" (database file);
"FMP Acknowledgments.pdf" file;
"Extensions" folder, containing a "Dictionaries" folder,
with the dictionary file for supported languages;
(and an "English" folder with 3 files, for Windows);
40 ".dll" files (for Windows).
- "calculate_bases.py" file (Python script which counts bases in a sequence file).
- DNA Folder with:
"DNA.app" (Macintosh) or "DNA.exe" (Windows) file
(the runtime application);
"DNA.fmp12" (database file);
"FMP Acknowledgments.pdf" file;
"Extensions" folder, containing a "Dictionaries" folder,
with the dictionary file for supported languages;
(and an "English" folder with 3 files, for Windows);
40 ".dll" files (for Windows).
- "calculate_bases.py" file (Python script which counts bases in a sequence file).
DNA
is based on FileMaker Pro 12 (FileMaker Pro, Inc.) database management
software (www.filemaker.com/index.html),
and is
released as a FileMaker Pro 12 template, along with a runtime
application able to run "FileMaker Pro" at the core of the
software.
The runtime is freely distributed, in compliance with the license of "FileMaker Pro 12 Advanced" developer package that was used to create the program.
The runtime is freely distributed, in compliance with the license of "FileMaker Pro 12 Advanced" developer package that was used to create the program.
Standard database commands (Find, Sort, Export
records) are available within each layout of DNA
(see "GENERAL DEFINITIONS"
and "MENU AND COMMANDS" sections in TGCA
software Guide).
Please do not change the names of any files and in the DNA folder.
Please do not change the names of any files and in the DNA folder.
NOTE - Be sure that
your system default format uses
"." (full stop)
as a decimal separator (English standard).
If this is not the case, you must change the system setting.
"." (full stop)
as a decimal separator (English standard).
If this is not the case, you must change the system setting.
Mac OS X: in
"System Preferences" (from the "Apple" Menu), click on
"International", then on "Formats", then choose as
"Region" a country with the English standard format for
numbers (full stop mark as a decimal separator).
System
restart or user logout is not required to make the
change effective.
Windows: in "Control Panel" (from the "Start" Menu), click on "International options" then modify the format of numbers choosing a country with the English standard format for numbers (full stop mark as a decimal separator).
System restart or user logout is not required to make the change effective.
Python 2.6 or 2.7 (https://www.python.org/) is only required to run the scripts useful for base counts.
Windows: in "Control Panel" (from the "Start" Menu), click on "International options" then modify the format of numbers choosing a country with the English standard format for numbers (full stop mark as a decimal separator).
System restart or user logout is not required to make the change effective.
Python 2.6 or 2.7 (https://www.python.org/) is only required to run the scripts useful for base counts.
1) Downloading chromosome sequences
Create a text file named "Chr_accession.txt" with a chromosome accessions list (an example can be downloaded by clicking here).
To download
chromosome sequences listed in the file named
"Chr_accession.txt" go to the website page:
http://www.ncbi.nlm.nih.gov/sites/batchentrez
http://www.ncbi.nlm.nih.gov/sites/batchentrez
On the web page select:
Database: Nucleotide;
File: click on the "Browse" button and select the "Chr_accession.txt" created
in the previous step.
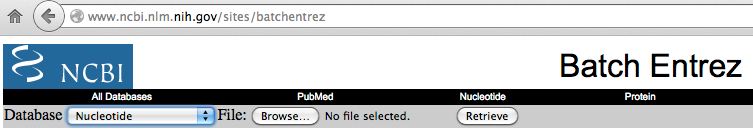
By clicking on the "Retrieve" button, a window with
the description of the retrieved records will appear.
Click on the "Retrieve
records for XXX UID(s)" link (where XXX is
the number of the retrieved records that should be
equal to the number of chromosome accessions listed in
the uploaded file). You can download the found entry set
choosing from the "Send to" pop-up menu at the
right-top corner of the web page:
"File", "FASTA" and "Default Order"; then clicking on the "Create File" button.
"File", "FASTA" and "Default Order"; then clicking on the "Create File" button.
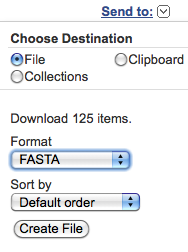
The download could
take some hours, depending on the number and the
size of chromosomes.
Please note that you should not exceed the download limit of 10 GB to avoid errors in the output file. You can divide the chromosome accession list file (chr_accession.txt) into two or more files and repeat the download step for each of them.
In the default download folder of your browser, you will obtain one or more files usually named "sequence.fasta", "sequence(1).fasta", "sequence(2).fasta" and so forth.
We recommend checking that the number of downloaded entries is equal to initial retrieved chromosome accession number (e.g. using the "grep" and "wc" UNIX utilities: grep gi sequence.fasta | wc -l).
Create a text file named exactly "file_list.txt" with a list of the downloaded FASTA file names (one row each name, even if you have only one file with the chromosome sequences, write only that name as in this example file).
Example
If you have only one file with the chromosome sequences, write only: sequence.fasta.
If you have three files with the chromosome sequences, write:
sequence.fasta
sequence(1).fasta
sequence(2).fasta
For Windows users only: you need to convert the FASTA file(s) in tabular format, for example by using Galaxy web tool.
Please note that you should not exceed the download limit of 10 GB to avoid errors in the output file. You can divide the chromosome accession list file (chr_accession.txt) into two or more files and repeat the download step for each of them.
In the default download folder of your browser, you will obtain one or more files usually named "sequence.fasta", "sequence(1).fasta", "sequence(2).fasta" and so forth.
We recommend checking that the number of downloaded entries is equal to initial retrieved chromosome accession number (e.g. using the "grep" and "wc" UNIX utilities: grep gi sequence.fasta | wc -l).
Create a text file named exactly "file_list.txt" with a list of the downloaded FASTA file names (one row each name, even if you have only one file with the chromosome sequences, write only that name as in this example file).
Example
If you have only one file with the chromosome sequences, write only: sequence.fasta.
If you have three files with the chromosome sequences, write:
sequence.fasta
sequence(1).fasta
sequence(2).fasta
For Windows users only: you need to convert the FASTA file(s) in tabular format, for example by using Galaxy web tool.
The "calculate_bases.py" Python script
provided here automatically counts all
types of nucleotide letters in
chromosome sequences.
You need to have the following files in the same folder:
1) the "calculate_sequences.py" script;
2) the FASTA file(s) with the chromosome sequences downloaded in section 1)
or the tabular format file of the chromosome sequences for Windows users only;
3) the text file "file_list.txt" with the list of the downloaded FASTA file names created in section 1).
Execute the "calculate_bases.py" script by typing the UNIX command "python calculate_sequences.py" or by running the script from the IDLE utility.
For those not used to UNIX and Python languages, we recommend using the IDLE utility to run Python scripts provided here. Please see the following quick guide (section B).
The programme is finished when you will obtain a file named "base_counts_2017.txt".
You need to have the following files in the same folder:
1) the "calculate_sequences.py" script;
2) the FASTA file(s) with the chromosome sequences downloaded in section 1)
or the tabular format file of the chromosome sequences for Windows users only;
3) the text file "file_list.txt" with the list of the downloaded FASTA file names created in section 1).
Execute the "calculate_bases.py" script by typing the UNIX command "python calculate_sequences.py" or by running the script from the IDLE utility.
For those not used to UNIX and Python languages, we recommend using the IDLE utility to run Python scripts provided here. Please see the following quick guide (section B).
The programme is finished when you will obtain a file named "base_counts_2017.txt".
3) Importing base counts
If the file created
in the previous step ("base_counts_2017.txt")
exceeds the size of 4 GB, please divide it in order
to create more files of less than 4 GB,
which is the size limit of text files to be imported
into a FileMaker database. Please repeat for each
file created
the following import step.
Open the DNA database by double-clicking on the DNA.app or DNA.exe icon.
Import the base counts by selecting the script "Import_Counts" from the GeneBase "Scripts" menu.
Open the DNA database by double-clicking on the DNA.app or DNA.exe icon.
Import the base counts by selecting the script "Import_Counts" from the GeneBase "Scripts" menu.
The DNA database will
calculate chromosome lengths and weights in specific calculated fields of
the "DNA"
table:
FIELD DESCRIPTION
"Chr":
the chromosome (chr) number;
"bp": the total of base pairs (bp) counted in each chr sequence; the sum
of this value for all the records is available at the bottom of the
window in the "Tot_bp" field;
"cm": the chromosome length calculated in centimeters (cm); the sum of
this value for all the records is available at the bottom of the
window in the "Tot_cm" field;
"pg": the chromosome length calculated in picograms (pg); the sum of this
value for all the records is available at the bottom of the window
in the "Tot_pg" field;
"Mbp": the total of Megabase pairs (Mbp) counted in each chr sequence; the
sum of this value for all the records is available at the bottom of
the window in the "Tot_Mbp" field;
"A": the sum of all the adenine (A) counted in each chr sequence;
"C": the sum of all the cytosine (C) counted in each chr sequence;
"T": the sum of all the thymine (T) counted in each chr sequence;
"G": the sum of all the guanine (G) counted in each chr sequence;
"bp": the total of base pairs (bp) counted in each chr sequence; the sum
of this value for all the records is available at the bottom of the
window in the "Tot_bp" field;
"cm": the chromosome length calculated in centimeters (cm); the sum of
this value for all the records is available at the bottom of the
window in the "Tot_cm" field;
"pg": the chromosome length calculated in picograms (pg); the sum of this
value for all the records is available at the bottom of the window
in the "Tot_pg" field;
"Mbp": the total of Megabase pairs (Mbp) counted in each chr sequence; the
sum of this value for all the records is available at the bottom of
the window in the "Tot_Mbp" field;
"A": the sum of all the adenine (A) counted in each chr sequence;
"C": the sum of all the cytosine (C) counted in each chr sequence;
"T": the sum of all the thymine (T) counted in each chr sequence;
"G": the sum of all the guanine (G) counted in each chr sequence;
"N":
the sum of all the N
(which stands for any nucleotide) counted in
each chr sequence;
each chr sequence;
"ATW": the sum of all the A, T and W (which stands for A or T) counted in
each chr sequence; the sum of this value for all the records is
available at the bottom of the window in the "Tot_ATW" field;
"GCS": the sum of all the G, C and S (which stands for G or C) counted in
each chr sequence; the sum of this value for all the records is
available at the bottom of the window in the "Tot_GCS" field;
"GCS%": the percentage of G, C and S calculated in the total of A, T, W, G,
C and S; the mean and the standard deviation for all the records are
shown in the "GCS%_Mean" and "GCS%_SD";
"ATW%": the percentage of A, T and W calculated in the total of A, T, W, G,
C and S; the mean and the standard deviation for all the records are
shown in the "ATW%_Mean" and "ATW%_SD";
"ATWGCS": the sum of all the A, T, W, G, C and S counted in each chr sequence;
the sum of this value for all the records is available at the bottom
of the window in the "Tot_ATWGCS" field;
"Uncertain_bp
bp-ATWGCS": the sum of uncertain bases, which is the difference between the
total of bp minus the total of A, T, W, G, C and S; the sum of this
value for all the records is available at the bottom of the window
in the "Tot_Uncertain" field;
"%": the percentage of uncertain bases;
"Uncertain
ATW_Estimation": the A, T and W composition of uncertain bases proportionately
estimated using the ATW sum; the sum of this value for all the
records is available at the bottom of the window in the
"Tot_Uncertain ATW_Estimation" field;
"Uncertain
GCS_Estimation": the G, C and S composition of uncertain bases proportionately
estimated using the GCS sum; the sum of this value for all the
records is available at the bottom of the window in the
"Tot_Uncertain GCS_Estimation" field;
"Uncertain
GCS%_Estimation": the percentage of the estimated G, C and S in the uncertain bases;
this value is actually equal to the GCS% value since the uncertain
base composition has been proportionately estimated and it is used
as an internal control;
"GCS%
bpTot": the percentage of the sum of the counted G, C, and S in certain
bases and the estimated G, C and S in the uncertain bases; this
value is actually equal to the GCS% value since the uncertain
base composition has been proportionately estimated and it is used
as an internal control;
"ATW%
bpTot": the percentage of the sum of the counted A, T, and W in certain
bases and the estimated A, T and W in the uncertain bases; this
value is actually equal to the ATW% value since the uncertain
base composition has been proportionately estimated and it is used
as an internal control;
"R": the sum of all the R (which stands for A or G) counted in each chr
sequence;
"S": the sum of all the S (which stands for G or C) counted in each chr
sequence;
"W": the sum of all the W (which stands for A or T) counted in each chr
sequence;
"M": the sum of all the M (which stands for A or C) counted in each chr
sequence;
"V": the sum of all the V (which stands for A, C or G) counted in each
chr sequence;
"H": the sum of all the H (which stands for A, C or T) counted in each
chr sequence;
"Y": the sum of all the Y (which stands for C or T) counted in each chr
sequence;
"K": the sum of all the K (which stands for G or T) counted in each chr
sequence;
"B": the sum of all the B (which stands for C, G or T) counted in each
chr sequence;
"D": the sum of all the D (which stands for A, G or T) counted in each
chr sequence;
"Accession": the chr accession number.