2.0 version (2011)
Definition
'5'_ORF_Extender' is a tool that can perform a systematic identification of extended coding regions (CDS) at the 5´ end of known mRNAs, using an EST-based approach.
Download
'5'_ORF_Extender' is a tool that can perform a systematic identification of extended coding regions (CDS) at the 5´ end of known mRNAs, using an EST-based approach.
Download
Download
Macintosh
version
(filled with human data and results)
Download Windows version (filled with human data and results)
Fast access to list of RefSeq human mRNAs not further extendable at their 5´ coding sequence (CDS) [RefSeq_mRNAs_with_Complete_ORF.Human.txt]
Fast access to list of RefSeq human mRNAs with an EST-based extension at their 5´ coding sequence (CDS) [Results.Human.xls]. These mRNAs correspond to 477 human loci [Loci.txt]
Download empty (template) Macintosh version
Download empty (template) Windows version
Download Windows version (filled with human data and results)
Fast access to list of RefSeq human mRNAs not further extendable at their 5´ coding sequence (CDS) [RefSeq_mRNAs_with_Complete_ORF.Human.txt]
Fast access to list of RefSeq human mRNAs with an EST-based extension at their 5´ coding sequence (CDS) [Results.Human.xls]. These mRNAs correspond to 477 human loci [Loci.txt]
Download empty (template) Macintosh version
Download empty (template) Windows version
Description of the main steps of the analysis
First, the user
is guided to import RefSeq mRNA
genomic coordinates
and sequences from UCSC genome browser.
In addition, a table, matching each mRNA or EST sequence of the investigated organism to a genomic locus, is imported following its obtainment from UniGene data.
In addition, a table, matching each mRNA or EST sequence of the investigated organism to a genomic locus, is imported following its obtainment from UniGene data.
At
the end of
this step, the software will have automatically determined
which mRNA
is candidate for extension of its 5´ CDS, due to the
absence
of an in-frame stop
codon upstream of the
described
initiation codon in the mRNA sequence entry.
Then, the user is guided to download the EST genomic coordinates from the UCSC Genome Browser.
The user will import these data for the EST assigned to all mRNAs potentially further extendable at their 5´ CDS.
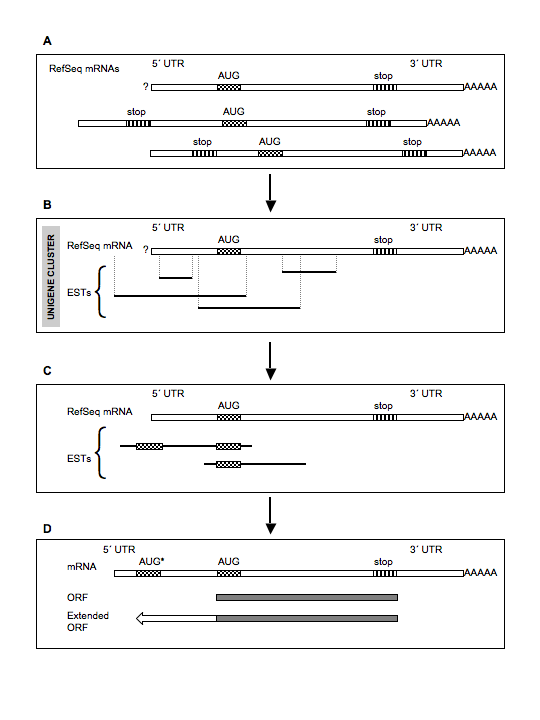
Following the download, from
the UCSC Genome Browser, of the sequences of the candidate
ESTs, the
software will determine which
EST
is suitable to extend its cognate mRNA 5´ CDS, due to the
presence
of a start codon upstream
the mRNA
known start codon and in frame with
it.
A sample screenshot of the main results is presented for QARS gene:

Database construction - Homo sapiens
Firstly, the human RefSeq flat file (version October 18, 2011) was downloaded from the UCSC (University of California, Santa Cruz) Genome Bioinformatics web site (http://genome.ucsc.edu/ - "Tables" section). The text file was imported into the appropriate 5'_ORF_Extender database table (following the software user guide) to obtain a local RefSeq database with 31,903 entries containing human known reference mRNA sequences ("NM_" prefix, thus excluding RefSeq entries not supported by experimental evidence, such as "XM_" models), corresponding to 18,665 distinct loci (a locus can have more than one mRNA isoform registered in RefSeq). It is possible to only select and further analyze mRNA entries without an in-frame stop codon upstream of the described initiation codon, which are thus candidates for a possible extension at 5´ end, because the presence of such a stop codon would indicate that the 5´ UTR sequence cannot be part of a longer continuous CDS. This also implies that a database of all RefSeq mRNAs that are bona fide complete at the 5´ end of their CDS is therefore generated.
The genome alignment data for human ESTs - assigned by UniGene (see section 2.3 below) to the same locus of the mRNAs candidates for a possible extension at 5´ end - were then downloaded from the UCSC site (7,166,113 entries, version October 19, 2011) and imported into the appropriate 5'_ORF_Extender table. Each human mRNA (without an in-frame stop codon upstream of the described initiation codon) was then compared with all the human EST assigned to the same locus by analyzing the coordinates of the pre-computed genome alignments for mRNAs and ESTs obtained by UCSC. Only those EST sequence entries presenting additional nucleotides upstream of the known 5´ mRNA end and therefore candidate to potentially extend the mRNA CDS at its 5´, were downloaded (159,378 candidates).
The whole analysis for H. sapiens, including data import and processing, required about 3 days for completion (1 additional day was required to obtain the updated UniGene table from 'UniGene Tabulator' and a further half day to import it prior to starting the analysis).
Tutorial
This Tutorial
guides
the user to a
step-by-step process in order to use
the software for the analysis of any organism.